Appendix: Obtaining a BED of Telomere sequences¶
Rationale¶
The Rhee and Pugh, 2011 paper used genomic features to filter out results from peak-calling. We want to do this as well.
A good approach seems:
- get list or lists as Bed file fom YeastMine
- Uses Bedtools with
intersectBed -vto filter out those summits that are in telomeres.
Obtain Telomeric sequences as a BED file from YeastMine¶
Go to YeastMine
Scroll down to where you see Lists.
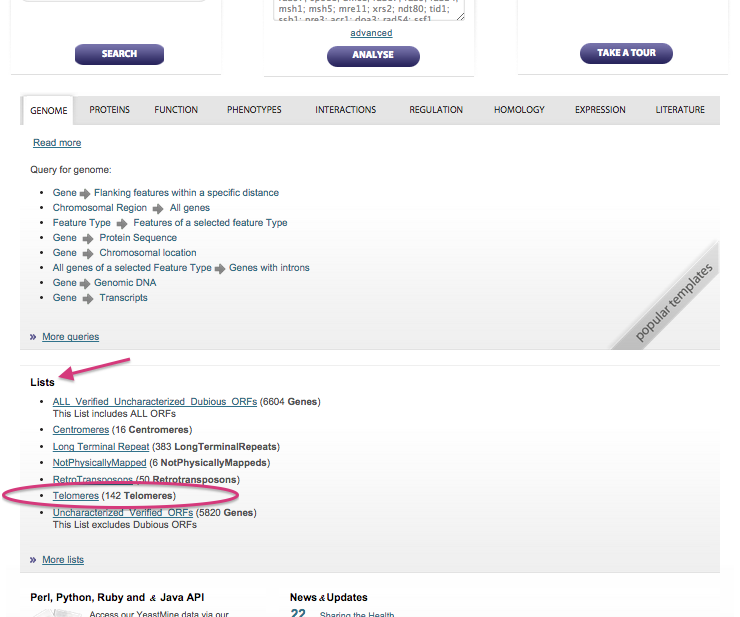
Among the Lists listed there should be Telomeres. If not, click
on More lists and examine the list of lists.
Click on the Telomeres list link.
Click on the Download button.
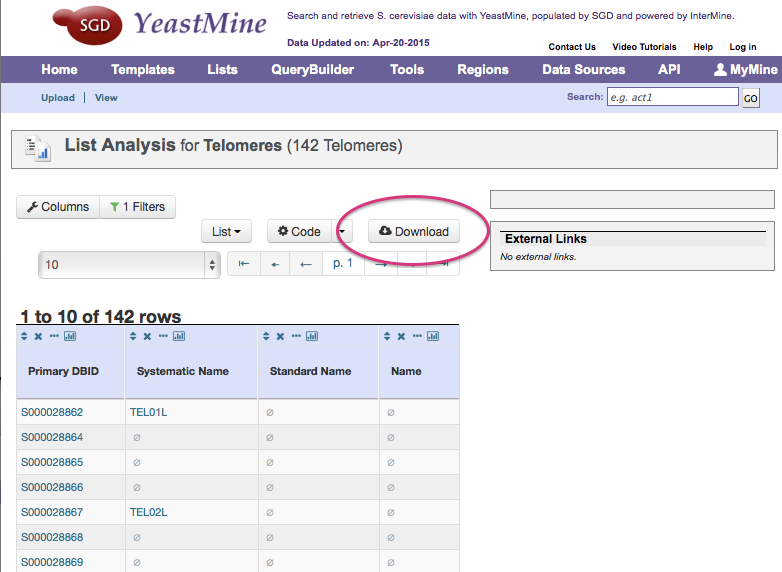
Choose UCSC-BED (Browser Extensible Display Format) from the
choices.
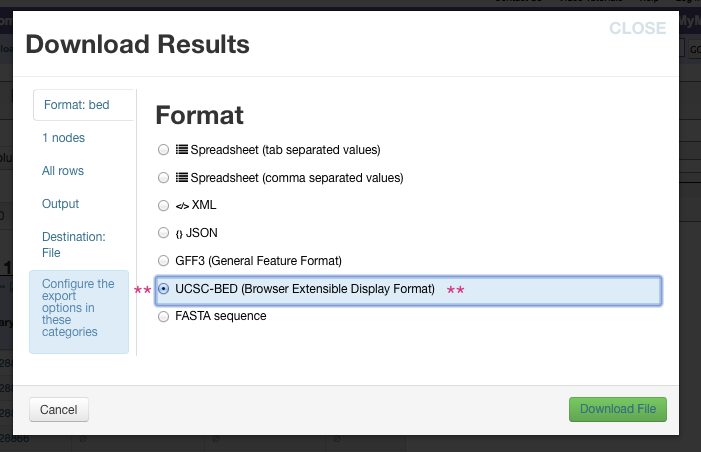
Now either click green Download File button at the bottom to
download to your local computer. You could then use scp to upload as
illustrated
here
it to an Amazon EC2 instance, for working on the worksop analysis
example.
Alternatively, click on the Destination File tab in the toolbar on
the left side of the Download Results window.
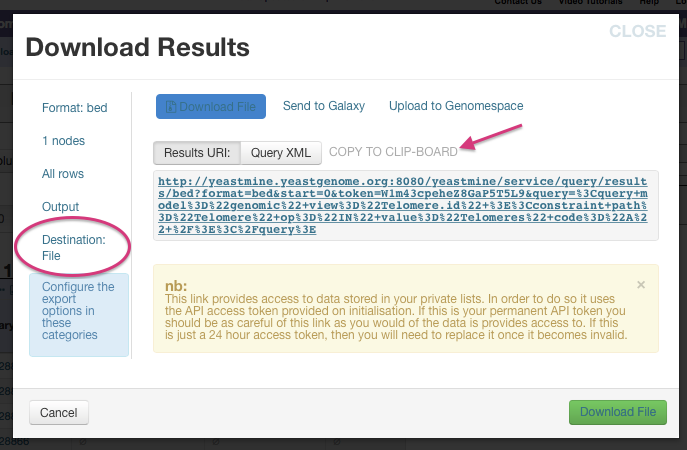
Note that there is a URL that makes uploading to an EC2 instance on
Amazon Web Services even easier. In fact you can click
copy to clipboard and then paste after the following text once you
ssh into your working directory on your instance. For example,
wget http://yeastmine.yeastgenome.org/yeastmine/service/query/results?format=tab&start=0&token=i1r4ud70h133R7ned6x2&columnheaders=1&query=%3Cquery+model%3D%22genomic%22+view%3D%22Telomere.primaryIdentifier+Telomere.secondaryIdentifier+Telomere.symbol+Telomere.name%22+%3E%3Cconstraint+path%3D%22Telomere%22+op%3D%22IN%22+value%3D%22Telomeres%22+code%3D%22A%22+%2F%3E%3C%2Fquery%3E
Note that the above command is just an example for constructing your command. The API token in the URL is temporary and expired long ago.